Introduction
HQStage is a lightweight tool for installing and using HQS software.
Want to learn more about what you can do with our software?
Explore our quantum simulation use cases or read through the use cases section in this documentation.
Try HQStage in the cloud
The fastest way to explore HQStage (and the full HQS software stack) is to run it directly in your browser on our managed JupyterLab service.
- Sign in at https://cloud.quantumsimulations.de
- Get free credits (
HQ$) at https://cloud.quantumsimulations.de/account/credits - Launch JupyterLab at https://notebook.cloud.quantumsimulations.de/
- Start using HQS modules with HQStage — no local setup required. We have pre-installed all eligible HQS Modules and common packages like numpy into your Python environment.
- Explore our use-cases and examples — downloaded and ready to use in your Notebooks
- You can start on our free plan; see Licensing for details
Code for NMR example
Code for NMR example
Copy and paste the following code into your first jupyter notebook to produce the plot shown in above screenshot
from hqs_nmr_parameters import examples
from hqs_nmr import NMRCalculationParameters, calculate_spectrum
from matplotlib import pyplot as plt
print(f"Available moleucules: {examples.molecules.keys}")
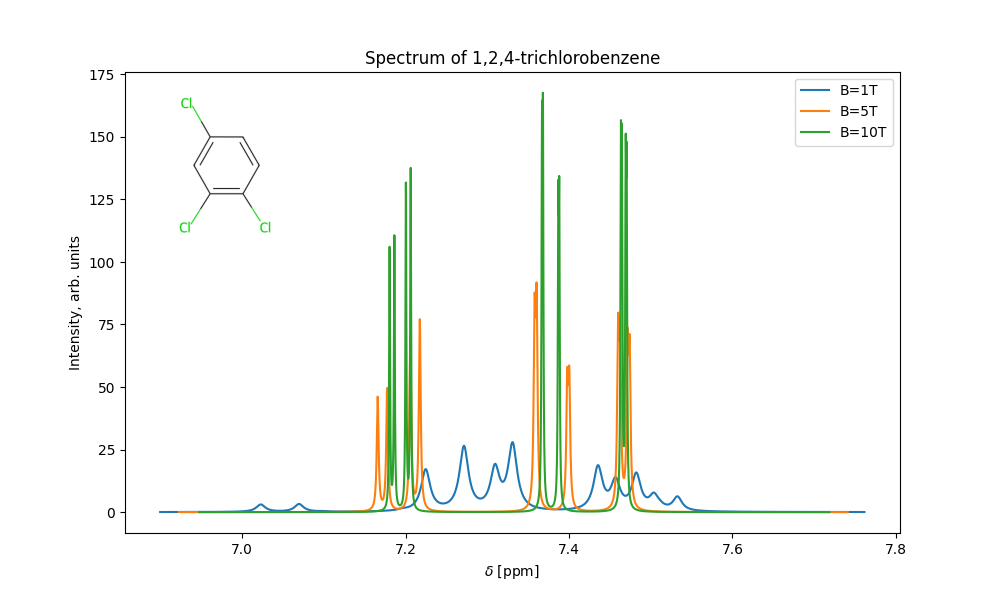
molecule_name = '1,2,4-trichlorobenzene'
molecule_parameters = examples.molecules[molecule_name]
fig, ax = plt.subplots(figsize=(10, 6))
for field_T in [1, 5, 10]:
result = calculate_spectrum(molecule_parameters, NMRCalculationParameters(field_T=field_T))
ax.plot(result.spectrum.omegas_ppm, result.spectrum.intensity, label=f"B={field_T}T")
plt.legend()
ax.set_xlabel(r"$\delta$ [ppm]")
ax.set_ylabel("Intensity, arb. units")
ax.set_title(f"Spectrum of {molecule_name}")
plt.show()
After starting your server your HQ$ balance will be debited until you shutdown the server.
After 6 hours of inactivety we will automatically shut down your server and stop debiting your HQ$ balance.
To shutdown the server click File-->Hub Control Panel-->Stop My Server. This will shutdown your server
and your HQ$ balance will no longer be debited.
Local Usage
Prerequisites
- Python 3.10+ (use the
uvtabs to install Python on the fly) - macOS/Linux
- Account on https://cloud.quantumsimulations.de
- Get a free trial license at https://cloud.quantumsimulations.de/licenses
- Proceed with installation
Installation
While it’s best to start in a fresh virtual environment, you can simply pip install HQStage into any existing Python environment. Below we show both options and also our preferred way of using HQStage with uv.
pip install hqstage
hqstage init
The hqstage init command will prompt you to create an account and user token on https://cloud.quantumsimulations.de.
Intel’s Math Kernel Library
Intel’s Math Kernel Library
Some HQStage Modules use Intel’s MKL library to accelerate mathematical calculations. You can install MKL using
hqstage install mkl
The command ensures that runtime libraries of mkl are present in the correct format in your environments lib folder.
Free trial
To get started with your first quantum computing use case request a free trial. This free trial will add the HQS Spectrum Tools to your account which we will use to run our first example use case: a NMR spectrum prediction.
Running your first use case example: NMR
Install the requried modules
hqstage install ipython jupyter matplotlib mkl hqs_spectrum_tools
and open an ipython interpreter:
ipython
Now, copy paste the following code snippet:
from hqs_nmr_parameters import examples
from hqs_nmr import NMRCalculationParameters, calculate_spectrum
from matplotlib import pyplot as plt
from rdkit.Chem import Draw, MolFromMolBlock
print(f"Available moleucules: {examples.molecules.keys}")
molecule_name = '1,2,4-trichlorobenzene'
molecule_parameters = examples.molecules[molecule_name]
rdkit_mol = MolFromMolBlock(molecule_parameters.structures["Molfile"].content)
mol_image = Draw.MolToImage(rdkit_mol)
fig, ax = plt.subplots(figsize=(10, 6))
for field_T in [1, 5, 10]:
result = calculate_spectrum(molecule_parameters, NMRCalculationParameters(field_T=field_T))
ax.plot(result.spectrum.omegas_ppm, result.spectrum.intensity, label=f"B={field_T}T")
plt.legend()
ax.set_xlabel(r"$\delta$ [ppm]")
ax.set_ylabel("Intensity, arb. units")
ax.set_title(f"Spectrum of {molecule_name}")
ax_image = fig.add_axes(
[0.1,
0.6,
0.25,
0.25]
)
ax_image.imshow(mol_image)
ax_image.axis('off')
plt.show()
This will calculate the NMR spectrum for 1,2,4-trichlorobenzen and produce the following plot:
Downloading and using our examples
Download the use-case and spectrum examples
mkdir -p ~/HQStage/Examples
hqstage download-examples --download-dir ~/HQStage/Examples
jupyter lab --notebook-dir ~/HQStage/Examples
This will open jupyter lab in the folder where we have downladed the HQStage examples in. Fell free to explore the examples andadjust to your needs or continue reading more about our use cases
Use cases
The HQStage Modules are centered around solving quantum mechanical systems. HQStage provides access to a wide range of example systems and Hamiltonians. There is great interest in solving these systems and Hamiltonains on quantum computers.
HQStage examples and modules will be available for three use cases:
- Nuclear magnetic resonance (HQS Spectrum Tools)
- Electron spectroscopy (HQS Quantum Solver)
- Magnetic response
As of now we have published the nuclear magnetic resonance (NMR) and the Electron spectroscopy use case. Magnetic resonance use cases will be published later in 2025.
Nuclear magnetic resonance
Expert users in NMR spectroscopy should use our HQStage modules HQS Spectrum Tools.
To get a feeling for NMR spectroscopy try out our end-to-end
product HQSpectrum.
See here for an example on how to use HQS Spectrum Tools NMR Hamiltonians with Qiskit to simulate NMR on a quantum computer.
In the following we will naswer the question why NMR is a good use case for quantum computers. For this we use our ITBQ use case evaluation.
I. Identify industry problem. NMR is a routine, cornerstone analytical technique for chemistry and pharma, used to determine molecular structure and to quantify mixtures (qNMR). Spectra encode structure through chemical shifts and J‑couplings, which depend on molecular environment (solvent, temperature, pH, field strength) and can be complex, especially for 1H spectra and at lower magnetic fields (benchtop devices).
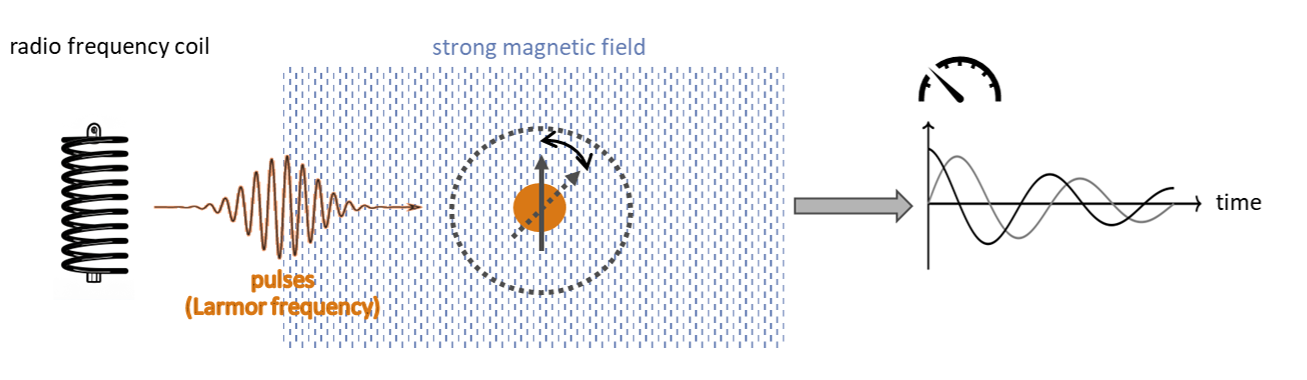
II. Transform to quantum. In the “transform to quantum” step we map molecules onto a quantum-mechanical spin model in which each nucleus is a spin-1/2 in a static magnetic field, and the spectrum is governed by Zeeman terms corrected by chemical shifts and by scalar J-couplings between spins; for diamagnetic molecules in liquids, direct dipolar couplings average out with molecular motion, while for macromolecules/solids they can matter. The resulting model is the Spin Hamiltonian:
Operationally, the transformation proceeds by generating 3D geometries and relevant conformers from molecular structure, optimizing them, computing shieldings and J-couplings (e.g., via DFT), Boltzmann-averaging parameters over the conformer ensemble, and converting shieldings to chemical shifts for use in the Hamiltonian; while these procedures are established and largely automated, integration into analysis workflows and accurate treatment of environmental effects still require manual checks. In HQS Spectrum Tools we provide a collection of pre-calculated spin models for a range of molecules with different size and chemical structures. Use these as a starting point for your NMR calculations - classical or on quantum computers. Rating: ★★★★☆.
III. Get the job done without the quantum computer.
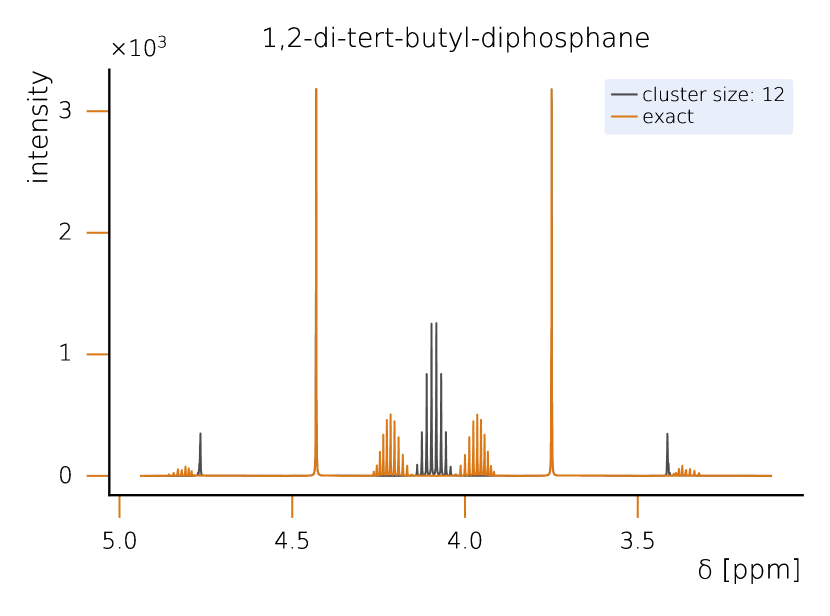
This section sets a clear classical baseline for quantum advantage in NMR simulations. Exact NMR dynamics scales exponentially with the number of spins, but with HQS Spectrum Tools practical, accurate results are routinely achieved by combining spin‑clustering (grouping strongly interacting spins) with symmetry reductions (e.g., local SU(2)), as implemented in the HQS Spectrum Tools and benchmarked against Spinach. Under typical high‑field conditions, these methods compute 1H spectra for organic molecules with tens of protons in minutes; for example, a 50‑spin Friedelin spectrum converges with clusters of about 8–12 spins. When correlations extend across the whole molecule, simple clustering can miss key features, yet symmetry‑aware exact modes still reproduce spectra for medium‑size systems, as shown for the 22‑spin 1,2‑di‑tert‑butyl‑diphosphane.
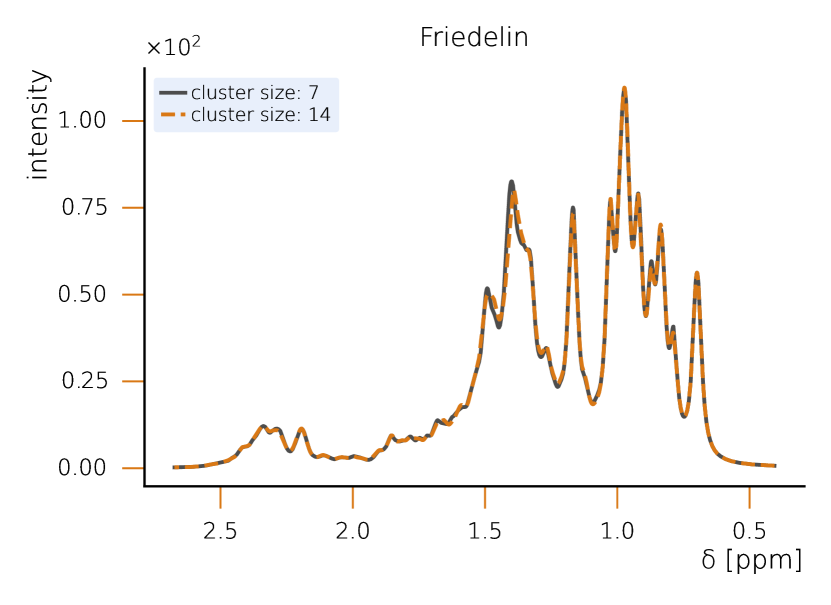

IV. NMR spectral prediction reduces to tracking the time‑dependent transverse magnetization of an N‑spin system, expressed as
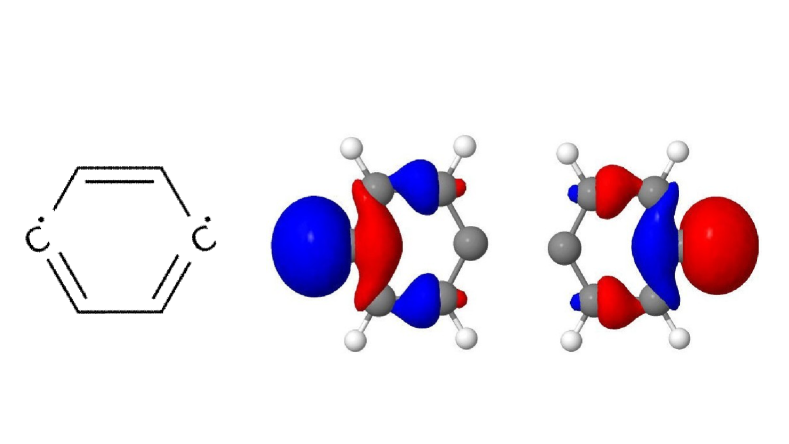
While quantum computing offers an exponential advantage for NMR simulations, classical algorithms have also been developed to efficiently treat large spin systems. One particularly effective approach is spin clustering (see Fig. (2)), which reduces computational complexity by grouping strongly interacting spins together, enabling approximate solutions. In many cases, especially at high magnetic fields, this method can provide accurate results with cluster sizes of 8–12 spins, even for complex molecules. For example, in the case of Friedelin (Fig. 2), a 50-spin molecule, the cluster-based solver shows good convergence, with minimal spectral changes as the cluster size increases. This demonstrates that for certain systems with relatively weak spin correlations, spin clustering provides a feasible classical approach.
However, when strong correlations extend across the system, as in the case of 1,2-di-tert-butyl-diphosphane (Fig. 2), the limitations of spin clustering become apparent. This molecule consists of 22 coupled spins, where cluster approximations cannot fully capture the spin interactions. Instead, solving the system exactly requires the exploitation of local SU(2) symmetry, which significantly reduces the computational complexity while maintaining full accuracy. Without symmetry considerations, the problem would require the diagonalization of a -dimensional matrix, making classical exact diagonalization basically intractable. Given examples like 1,2-di-tert-butyl-diphosphane, it seams likely that parameters example and parameter regimes exist in which classical solutions remain challenging. At the same time it is clear that classical solvers can cover a large part of the relevant cases for NMR. Therefore we rate the probability to achieve quantum advantage with four stars (★★★★★).
Electron Spectroscopy (Singlet–Triplet Splitting)
Electron spectroscopy measures excited states of molecules. Depending on the setup, it can probe purely electronic excitations, vibrational structure on top of them, or their coupling. A particularly important quantity that these measurements access is the singlet–triplet energy splitting ΔEST between the lowest singlet (S1) and triplet (T1) states. In what follows we focus on accurately predicting ΔEST for realistic molecules.
I. Identify industry problem As an example for the importance of ΔEST we will discuss OLEDs. In commercial OLED displays and phones, the light‑emitting molecule (the emitter) is dissolved at low concentration in an organic‑semiconductor host. Its emission comes from the emitter’s lowest triplet state T1, so we must know where that level sits. The singlet–triplet splitting ΔEST fixes T1 once the singlet S1 is known, making accurate ΔEST a design‑critical number. For a stable, efficient device the emitter’s T1 must be lower than the triplet energies of the host and neighboring layers—i.e., not resonant with the host’s triplet manifold. If T1 is too close to, or above, the host triplet level, triplet excitons can hop from the emitter to the host via short‑range energy transfer or form interfacial charge‑transfer states. On the host these excitations are usually lost non‑radiatively, and they can also react with charge carriers or impurities to break chemical bonds. The outcomes are poorer efficiency, worse color purity and shorter lifetime. Hence, device engineers need reliable ΔEST predictions to place the emitter’s triplet safely below the host’s triplet energies. Similarly to OLEDs for displays we need ΔEST for other light emitting or absorbing materials.
II. Transform to quantum We start by building a sensible orbital basis for the molecule using Hartree–Fock (HF) or a very small CASSCF calculation to capture any obvious static correlation. From this basis we define an active space containing only the two frontier orbitals that control the singlet–triplet gap (two electrons in two orbitals). All remaining orbitals are “downfolded” with a random‑phase‑approximation (RPA) treatment: their collective response is represented as a set of effective harmonic resonators that screen and couple to the active space. This yields a compact effective Hamiltonian with a 4×4 electronic block (the two‑orbital problem) coupled to a handful of oscillators—i.e., a “two‑qubit plus oscillators” model. On a quantum computer the 4×4 block maps to two qubits and requires very shallow circuits; accuracy is dialed in by how many RPA modes we keep and by the truncation chosen for each mode. We then read out the singlet–triplet splitting ΔEST from this model (e.g., via energy estimation or shadow‑tomography style procedures), which will be discussed later in the quantum‑advantage section.
III. Get the job done without the quantum computer Our classical baseline is a straightforward multireference workflow built on PySCF with the Block DMRG solver. After preparing molecular geometries, we use the Active Space Finder (ASF) to choose the most important orbitals that control the singlet–triplet gap. The ASF is an open source tool developed by HQS in cooperation with Covestro and runs quick reference calculations, a DMRG calculation with low precision, and then proposes the smallest consistent set of “frontier” orbitals needed to form a consistent active space. This avoids trial‑and‑error and the need for manual expert guesses. With that active space fixed, we compute singlet and triplet energies using CASSCF; if more orbitals are needed for an accurate description, we swap the active‑space solver to DMRG via Block so we can treat larger spaces with the same overall setup. Finally, we include dynamical correlation on top of this multireference description using NEVPT2 or CASPT2. A major advantage as compared to our quantum computing approach is that CAS Methods are also routinely used to calculate various other energy difference of electronic states, which are also in principle accessible via electron spectroscopy. For more information about the ASF, read our paper.
IV. Show quantum advantage The RPA‑style reduction to a 4×4 electronic block coupled to a modest set of oscillators turns this into a problem whose quantum cost grows mainly with the number and truncation of those modes. So the problem is quite similar to the well known spin-boson problem, with a slightly enlarged spin system. Each boson can be in the cases we studied be mapped to a single qubit, since the RPA approximation depends on the occupation number of the bosons to stay low. But what is particularly interesting, is that the fact that we consider non-interacting boson, allows a large amount of mapping between the problem and the processor. It is well known in the study of spin boson systems, that the boson can be mapped to a 1-dimensional chain of bosons. This would allow simulation with constant depth even if the number of bosons increases ever further. Also many other mappings might be possible. Since we are looking at energy difference, it is also possible to extract the relevant quantities from time evolution under the system Hamiltonian. Here again we play towards the strength of quantum computing and can avoid e.g. variational approaches. A method like shadow spectroscpy should be sufficent to access all relevant results.
HQS Spectrum Tools
You can find the detailed User Documentation here.
HQS Quantum Solver
You can find the detailed User Documentation here.
Licensing & Modules
HQStage is an execution environment that can be enriched with a variety of modules. These can be obtained either by loan (test license), purchase (“Academic user/ node locked” licenses) or rental (“Industry user/ node locked” licenses) via download.
A list of the available modules can be found on the modules page where descriptions of the modules are also available. The corresponding licenses can be managed on the HQS Cloud.
As a rule, HQStage modules run on 64-bit systems with current Linux and Windows operating systems and also on current MacOS operating systems on ARM architectures. Please refer to the description or documentation of the modules for any restrictions on the execution environment.
HQS uses open source components (OSS components). These OSS components are listed in the respective module overview page in this documentation, along with the applicable license for the OSS components.
Within HQStage, the corresponding modules can be installed and updated based on the licenses available to the user. To learn more look at our local usage.
The following pages provide an overview of available HQStage modules and different license types.
Licensing
HQStage is an execution environment that can be enriched with a variety of modules. These can be obtained either by loan (test license), purchase (“Academic user/ node locked” licenses) or rental (“Industry user/ node locked” licenses) via download.
A list of the available modules can be found on the modules page where descriptions of the modules are also available. The corresponding licenses can be managed on the HQS Cloud.
As a rule, HQStage modules run on 64-bit systems with current Linux and Windows operating systems and also on current MacOS operating systems on ARM architectures. Please refer to the description or documentation of the modules for any restrictions on the execution environment.
HQS uses open source components (OSS components). These OSS components are listed in the respective module overview page in this documentation, along with the applicable license for the OSS components.
Within HQStage, the corresponding modules can be installed and updated based on the licenses available to the user. To learn more look at our local usage section.
The following pages provide an overview of available HQStage license types.
Obtaining a license
In order to obtain and manage HQStage licenses you have to create an account on the HQS Cloud.
All licenses are bound to our General Terms and Conditions. This General Terms and Conditions always take precedence over this documentation. The following list of available licenses provides only a summary of the licenses defined in the General Terms and Conditions.
When running HQStage and its modules the validity and scope of your licenses will be verified, this requires internet access of the respective machine.
Available Plans
We offer four plans for HQStage and HQS Cloud. A free (trial) plan, a standard (cloud only) plan and premium plans for academic and commercial users. Buying or subscribing to HQStage Modules unlocks the premium plan. See the following table for an overview of the different plans:
| Plan | Free | Standard | Premium (Academic) | Premium (Commercial) |
|---|---|---|---|---|
| HQStage | free trial license. | download of Modules not possible (Cloud only) | purchase selected HQStage Modules (*) | subsribe to selected HQStage Modules (*) |
| download HQStage | ✅ | ❌ | ✅ | ✅ |
| run modules locally | ✅ | ❌ | ✅ | ✅ |
| full control over hardware and data | ✅ | ❌ | ✅ | ✅ |
| use in your own toolchain | ✅ | ❌ | ✅ | ✅ |
| integrate with your own libraries | ✅ | ❌ | ✅ | ✅ |
| downloadable modules | ✅ HQS Noise App Free ✅ HQS Qorrelator App Free | ❌ | ✅ purchased Modules | ✅ subscribed Modules |
| software updates | during free period | ❌ | ✅ for one year after purchase | ✅ During active subscription |
| HQS Cloud | Try out all modules with up to 10 HQ$ as free credits. | Get cloud access to all modules. Buy computing credits and pay as you go on HQS Cloud. | Purchase of HQStage Modules unlocks cloud access to all modules. Buy computing credits and pay as you go on HQS Cloud. | Subscription to HQStage Modules unlocks cloud access to all modules. Buy computing credits and pay as you go on HQS Cloud. |
| use HQStage inside your browser | ✅ | ✅ | ✅ | ✅ |
| access to all modules | ✅ | ✅ | ✅ | ✅ |
| unlimited qubits (**) | ❌ | ✅ | ✅ | ✅ |
| install and use PyPI packages | ✅ | ✅ | ✅ | ✅ |
| use HQSTasks (HPC power) | ✅ | ✅ | ✅ | ✅ |
| free Computing credits | 10HQ$ (on request) | ❌ | 600HQ$$ per purchased module point | 50HQ$ per subscribed module point per month |
| buy additional credits | ❌ | 1HQ$ for 5€ | 1-3 Module points: 1HQ$ for 3€ 4-10 Module points: 1HQ$ for 2€ 10+ Module points: 1HQ$ for 1.5€ | 1-3 Module points: 1HQ$ for 3€ 4-10 Module points: 1HQ$ for 2€ 10+ Module points: 1HQ$ for 1.5€ |
| Territories | No restrictions. | access will only for customers in selected territories (***) | access will only for customers in selected territories (***) | access will only for customers in selected territories (***) |
To learn more about available modules and licensing visit the licensing and modules section of this documentation. The free trial license (free plan) can be requested here. To unlock a advanced plan, obtain a license or upgrade an existing license, please contact us.
(*) See here for module points and pricing
(**) The number of qubits in the HQS Noise App, the HQS Qorrelator App and the HQS Quantum Libraries are restricted in the Free plan.
To indicate the difference the Modules are named HQS Noise App Free, HQS Qorrelator App Free and HQS Quantum Libraries Free in this case.
(***) A list of available regions can be found here
Available Licenses
The free trial license includes access to HQStage with the following modules
and the following conditions apply
- Duration 6 months - after this period access to the modules will be withdrawn.
- The trial license is tied to a specific user and cannot be transferred.
- The license can only be activated on one machine at a time. The activated machine can be changed once. Machines can only be activated once for a free trial license. Other users cannot activate the free trial license on a previously activated machine.
You can request a free trial here.
Academic users and institutions
For academic users, HQS offers user-locked and node-locked licenses for purchase (see price list for prices). These licenses are linked to a specific user or one machine at a time respectively.
Users from the academic sector can purchase licenses for HQStage modules (see price list for prices), which are linked to specific users and are subject to the following conditions:
- During the first year after purchase, new versions of the Software, if any, will be provided.
- The license is bound to certain users. A change of activated users is possible on request. The users must belong to an academic institution (college, university, research institute).
- The license can be activated on any number of machines.
- Chargeable updates after the end of the first year and support are only possible by separate agreement.
Users from the academic sector can purchase licenses for HQStage modules (see price list for prices), which are linked to specific machines and are subject to the following conditions:
- During the first year after purchase, new versions of the Software, if any, will be provided.
- The license can be used by any number of users. The users must belong to an academic institution (college, university, research institute).
- The license can only be activated on one machine (node) at a time.
- Chargeable updates after the end of the first year and support are only possible by separate agreement.
Commercial users and institutions
HQS offers user-locked and node-locked subscription models for business and industrial users. For pricing, please request a quote.
Commercial users can purchase licenses for HQStage modules on a rental basis (see price list for prices), which are tied to specific users and are subject to the following conditions:
- Duration 1 year - after which access to modules will be withdrawn
- The license is bound to certain users. A change of activated users is possible on request.
- The license can be activated on any number of machines.
- New software versions, if any, will be provided during the term.
Commercial users can purchase licenses for HQStage modules on a rental basis (see price list for prices), which are linked to specific machines and are subject to the following conditions:
- Duration 1 year - after which access to modules will be withdrawn
- The license can be used by any number of users.
- The license can only be activated on one machine (node) at a time.
- New software versions, if any, will be provided during the term.
How to purchase a license
The free trial license can be requested here. To obtain another license or upgrade an existing license, please contact us.
Pricing
Academic users and institutions
HQStage uses a point system to indicate the size and complexity of the various Modules. The price depends on the total number of points for the modules selected and the number of users or machines required.
| Module Points | User-locked: Price per User | Node-locked: Price per Machine |
|---|---|---|
| 1 module point | 5,000.00 € | 5,000.00 € |
| 2 module points | 10,000.00 € | 10,000.00 € |
| 3 module points | 15,000.00 € | 15,000.00 € |
| 4 module points | 19,750.00 € | 19,750.00 € |
| 5 module points | 24,500.00 € | 24,500.00 € |
| 6 module points | 29,250.00 € | 29,250.00 € |
| 7 module points | 33,750.00 € | 33,750.00 € |
| 8 module points | 38,250.00 € | 38,250.00 € |
| 9 module points | 42,750.00 € | 42,750.00 € |
| per additional module point | 4,250.00 € | 4,250.00 € |
For larger numbers of users or machines, please contact us for a quote.
HQStage Modules have the following module points value:
| Application | Module Points |
|---|---|
| HQStage | 0 module points |
| HQS Noise App | 1 module point |
| HQS Qorrelator App | 1 module point |
| HQS Modeling Assistant | 1 module point |
| HQS Spin Mapper | 1 module point |
| HQS Molecules | 1 module point |
| HQS Quantum Libraries | 1 module point |
| HQS Quantum Solver | 1 module point |
| HQS Spectrum Tools | 1 module point |
| HQS Qolossal | 1 module point |
| HQS Raqet | 1 module point |
Commercial users and institutions
Prices available on request
Territorial restrictions
Some plans (Standard, Premium) are only available in the following countries / regions:
| European Union |
| US |
| Australia |
| Japan |
| Canada |
| New Zealand |
| Norway |
| UK |
| Switzerland incl Lichtenstein |
Modules
At HQS we believe that all steps of the simulation of quantum mechanical systems belong to one of four categories. All our available modules are organized along these categories:
Easy input
Easy input
When we design our tools, our focus is on creating clear and intuitive interfaces. This approach not only simplifies the use of each tool on its own, but also ensures they can be easily interconnected for more comprehensive tasks. Nonetheless, the inherent complexity associated with scientific software is undeniable. To mitigate this and enhance user experience, we provide the HQS Notebook Assistant. This AI-powered chat assistant simplifies the onboarding and use of our software, making it more accessible and user-friendly.
Model analysis
Model analysis
Before starting a simulation, you need to create a detailed description of the system you want to simulate. This description should be as accurate as possible and compatible with the simulation methods you’re using. With HQS Molecules, we make it easy to turn a molecule’s name into a 3D model on your computer. Also, our Spin Mapper tool lets you convert an electronic description into a spin-based one. This gives you more options for simulation methods and makes it possible to simulate these systems on quantum computers.
Quantum computer
Quantum computer
Quantum advantage has been shown for abstract problems, but practical benefits are still limited. HQS’s approach to quantum computing aims to address this by focusing on time evolutions on quantum computers and managing noise in a unique way. The HQS Noise App provides a new way to translate problems from fields like chemistry, materials science, and other quantum mechanical problems to a quantum computer. The HQS Qorrelator App is designed to calculate correlation functions, especially for NMR spectroscopy. With the HQS Quantum Libraries we offer a wide range of modules for advanced quantum computing tasks.
Simulation
Simulation
Discover the power of quantum simulation with HQS’ suite of simulation tools, designed to run on conventional computers. Study many-particle physics problems efficiently with the HQS Quantum Solver. Calculate spectra of spin systems with HQS Spectrum Tools in the context of NMR spectroscopy. Compute properties of materials and devices on a large scale with HQS Qolossal. Tackle the complexity of noisy quantum systems with HQS Raqet.
Available Modules
The following modules are available on HQStage. Following the links you can find the User Documentation of the Modules.
The customer is advised that HQS uses open source components (OSS components). These OSS components are listed in the respective module documentation, stating the applicable license of the OSS components.
To learn more about acquiring a license visit licensing.
HQS Spin Mapper
You can find the detailed User Documentation here.
HQS Molecules
You can find the detailed User Documentation here.
HQS Noise App
You can find the detailed User Documentation here.
HQS Qorrelator App
You can find the detailed User Documentation here.
HQS Quantum Libraries
The HQS Quantum Libraries encompass a comprehensive suite of modules tailored for advanced quantum computing tasks. These modules include algorithms, qonvert, noise-mapper, and the bath-mapper, serving to facilitate quantum algorithm development, compilation, noise emulation, and system-bath interaction modeling.
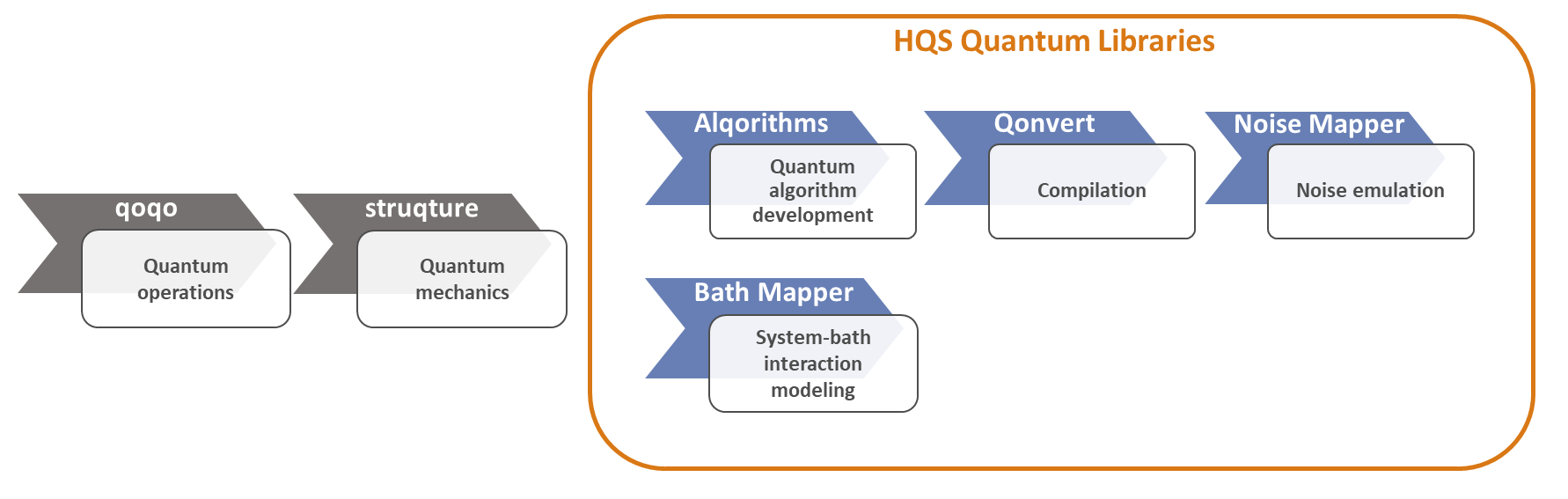
The HQS Quantum Libraries is a collection of software packages. These are as follows:
Alqorithms: a collection of tools for building quantum circuits. It includes a collection of quantum algorithms for simulating the trotterized time evolution of a quantum system, tools to build circuits for various kinds of measurements, and algorithms to compute two-point correlators. The detailed Documentation ofAlqorithms.Qonvert: a range of classes designed to “convert” circuits and quantum programs, each serving as a Converter with specific functionalities: gate decomposition, noise insertion, optimization and routing. The detailed Documentation ofQonvert.Noise Mapper: implements noise mapping, the construction of a static noisy algorithm model based on a quantum circuit and physical noise on a quantum computer. The detailed Documentation of theNoise Mapper.Bath Mapper: creates objects based on the Bloch-Redfield master equation and to manipulate them. The detailed Documentation of theBath Mapper.Bath Fitter: fits a given open quantum system with an effective open quantum system with finite degrees of freedom. The detailed Documentation of theBath Fitter.
The HQS Quantum Libraries need to be used in conjunction with the open-source HQS libraries qoqo and struqture, designed for creating quantum programs, as well as representing Hamiltonians and physical models.
Not sure where to start?
Take out quiz below, to see which example notebook best suits your needs and interests, and go from there!
HQS Qolossal
You can find the detailed User Documentation here.
HQS Raqet
You can find the detailed User Documentation here.
Restrictions and OSS
On this page we list
- Open source dependencies
- Installation hints
- System architecture / operating system restrictions
for all HQStage Modules.
Additionally, territorial restrictions apply.
HQS Spin Mapper
Details
OSS components installed by the user
OSS components installed by the user
Python packages
| Package | License | |
|---|---|---|
| 0 | Cerberus | ISC |
| 1 | PyYAML | MIT |
| 2 | annotated-types | MIT |
| 3 | asteval | MIT |
| 4 | contourpy | BSD License |
| 6 | cycler | BSD License |
| 7 | dill | BSD-3-Clause |
| 8 | fonttools | MIT |
| 10 | kiwisolver | BSD License |
| 14 | lmfit | BSD-3-Clause |
| 15 | matplotlib | PSF-2.0 |
| 16 | narwhals | MIT |
| 17 | numpy | BSD License |
| 18 | opt_einsum | MIT |
| 19 | packaging | Apache Software License |
| 20 | pillow | MIT-CMU |
| 21 | pip | MIT |
| 22 | plotly | MIT |
| 23 | pydantic | MIT |
| 24 | pydantic_core | MIT |
| 25 | pyparsing | MIT |
| 26 | python-dateutil | Apache Software License |
| 27 | qoqo_calculator_pyo3 | Apache-2.0 |
| 28 | scipy | BSD License |
| 29 | six | MIT |
| 30 | struqture-py | Apache-2.0 |
| 31 | types-PyYAML | Apache-2.0 |
| 32 | typing-inspection | MIT |
| 33 | typing_extensions | PSF-2.0 |
| 34 | uncertainties | BSD License |
HQS Molecules
Details
Please note the additional installation instructions for HQS Molecules in the documentation.
OSS components installed by the user
OSS components installed by the user
Python packages
| Package | License | |
|---|---|---|
| 0 | annotated-types | MIT |
| 1 | certifi | MPL-2.0 |
| 2 | charset-normalizer | MIT |
| 4 | idna | BSD-3-Clause |
| 5 | networkx | BSD License |
| 6 | numpy | BSD License |
| 7 | pip | MIT |
| 8 | pydantic | MIT |
| 9 | pydantic_core | MIT |
| 10 | requests | Apache-2.0 |
| 11 | scipy | BSD License |
| 12 | typing-inspection | MIT |
| 13 | typing_extensions | PSF-2.0 |
| 14 | urllib3 | MIT |
Additional binaries
| name | license |
|---|---|
| openbabel | GPL-2.0 license |
| xtb | LGPL-3.0 |
| crest | LGPL-3.0 |
HQS Noise App
HQS Qorrelator App
HQS Quantum Libraries
Details
Please note that the HQS Quantum Libraries are currently only supported on Linux and MacOS.
OSS components installed by the user
OSS components installed by the user
| Package | License | |
|---|---|---|
| 0 | annotated-types | MIT |
| 1 | asteval | MIT |
| 4 | build | MIT |
| 5 | dill | BSD-3-Clause |
| 7 | lmfit | BSD License |
| 9 | numpy | BSD License |
| 10 | packaging | Apache Software License |
| 11 | pip | MIT |
| 13 | pydantic | MIT |
| 14 | pydantic_core | MIT |
| 15 | pyproject_hooks | MIT |
| 17 | qoqo | Apache-2.0 |
| 18 | qoqo-quest | Apache-2.0 |
| 19 | qoqo_calculator_pyo3 | Apache-2.0 |
| 20 | scipy | BSD License |
| 21 | setuptools | MIT |
| 22 | struqture-py | Apache-2.0 |
| 23 | typing-inspection | MIT |
| 24 | typing_extensions | PSF-2.0 |
| 25 | uncertainties | BSD License |
Additional dependecy license files
Additional dependecy license files
QuEST_BSD_LICENSE_FILE Copyright (C) 1997 - 2002, Makoto Matsumoto and Takuji Nishimura, All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
1. Redistributions of source code must retain the above copyright
notice, this list of conditions and the following disclaimer.
2. Redistributions in binary form must reproduce the above copyright
notice, this list of conditions and the following disclaimer in the
documentation and/or other materials provided with the distribution.
3. The names of its contributors may not be used to endorse or promote
products derived from this software without specific prior written
permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT OWNER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE. QuEST_LICENSE_FILE MIT License
Copyright (c) 2017 aniabrown
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
HQS Quantum Solver
Details
Please note the additional installation instructions for MKL libraries.
OSS components installed by the user
OSS components installed by the user
Python packages
| Package | License | |
|---|---|---|
| 0 | annotated-types | MIT |
| 1 | attrs | MIT |
| 2 | contourpy | BSD License |
| 3 | cycler | BSD License |
| 4 | fonttools | MIT |
| 6 | kiwisolver | BSD License |
| 9 | matplotlib | PSF-2.0 |
| 10 | numpy | BSD License |
| 11 | opt_einsum | MIT |
| 12 | packaging | Apache Software License |
| 13 | pillow | MIT-CMU |
| 14 | pip | MIT |
| 15 | psutil | BSD-3-Clause |
| 16 | pydantic | MIT |
| 17 | pydantic_core | MIT |
| 18 | pyparsing | MIT |
| 19 | python-dateutil | Apache Software License |
| 20 | scipy | BSD License |
| 21 | six | MIT |
| 22 | sparse_dot_mkl | MIT |
| 23 | typing-inspection | MIT |
| 24 | typing_extensions | PSF-2.0 |
HQS Spectrum Tools
Details
Please note the additional installation instructions for MKL libraries.
OSS components installed by the user
OSS components installed by the user
Python packages
| Package | License | |
|---|---|---|
| 1 | PyYAML | MIT |
| 2 | annotated-types | MIT |
| 3 | contourpy | BSD License |
| 4 | cycler | BSD License |
| 5 | fonttools | MIT |
| 8 | kiwisolver | BSD License |
| 10 | matplotlib | PSF-2.0 |
| 11 | numpy | BSD License |
| 12 | opt_einsum | MIT |
| 13 | packaging | Apache Software License |
| 14 | pillow | MIT-CMU |
| 15 | pip | MIT |
| 16 | pydantic | MIT |
| 17 | pydantic_core | MIT |
| 18 | pyparsing | MIT |
| 19 | python-dateutil | Apache Software License |
| 20 | qoqo_calculator_pyo3 | Apache-2.0 |
| 21 | scipy | BSD License |
| 22 | setuptools | MIT |
| 23 | six | MIT |
| 24 | struqture-py | Apache-2.0 |
| 25 | types-PyYAML | Apache-2.0 |
| 26 | typing_extensions | PSF-2.0 |
HQS Qolossal
Details
Please note the additional installation instructions for MKL libraries.
OSS components installed by the user
OSS components installed by the user
Python packages
| Package | License | |
|---|---|---|
| 0 | Cerberus | ISC |
| 1 | PyYAML | MIT |
| 2 | annotated-types | MIT |
| 3 | asteval | MIT |
| 4 | contourpy | BSD License |
| 6 | cycler | BSD License |
| 7 | dill | BSD-3-Clause |
| 8 | fonttools | MIT |
| 9 | kiwisolver | BSD License |
| 13 | lmfit | BSD-3-Clause |
| 14 | matplotlib | PSF-2.0 |
| 15 | narwhals | MIT |
| 16 | numpy | BSD License |
| 17 | packaging | Apache Software License |
| 18 | pillow | MIT-CMU |
| 19 | pip | MIT |
| 20 | plotly | MIT |
| 21 | pydantic | MIT |
| 22 | pydantic_core | MIT |
| 23 | pyparsing | MIT |
| 24 | python-dateutil | Apache Software License |
| 26 | scipy | BSD License |
| 27 | setuptools | MIT |
| 28 | six | MIT |
| 29 | sparse_dot_mkl | MIT |
| 30 | typing-inspection | MIT |
| 31 | typing_extensions | PSF-2.0 |
| 32 | uncertainties | BSD License |